Including R in your Flat Data Workflow
Fetch and clean data automatically with GitHub Actions + R
May 2021 • 7 minute read
The GitHub OCTO team recently released their first project: Flat Data. The project aims to offer "a simple pattern for bringing working datasets into your repositories and versioning them." And it succeeds in doing so! I recently incorporated Flat Data into one of my projects, allowing me to finally stop manually updating the data on a semiregular basis (yikes!). While working, I couldn't find any documentation on using R with Flat Data. Here, I'll explain the steps I took to incorporate R scripts into a Flat Data pipeline.
Note: If you want to follow along, the GitHub repo can be found here.
What's Flat Data?
Flat Data solves the problem of carrying out the same repetitive tasks—retrieving, cleaning, and then republishing data—that commonly affects developers who want to present rapidly updating data (for example, COVID-19 data that updates daily). And although alternative solutions exist, Flat Data is easy, intuitive, and integrated directly with your GitHub repository (via GitHub):
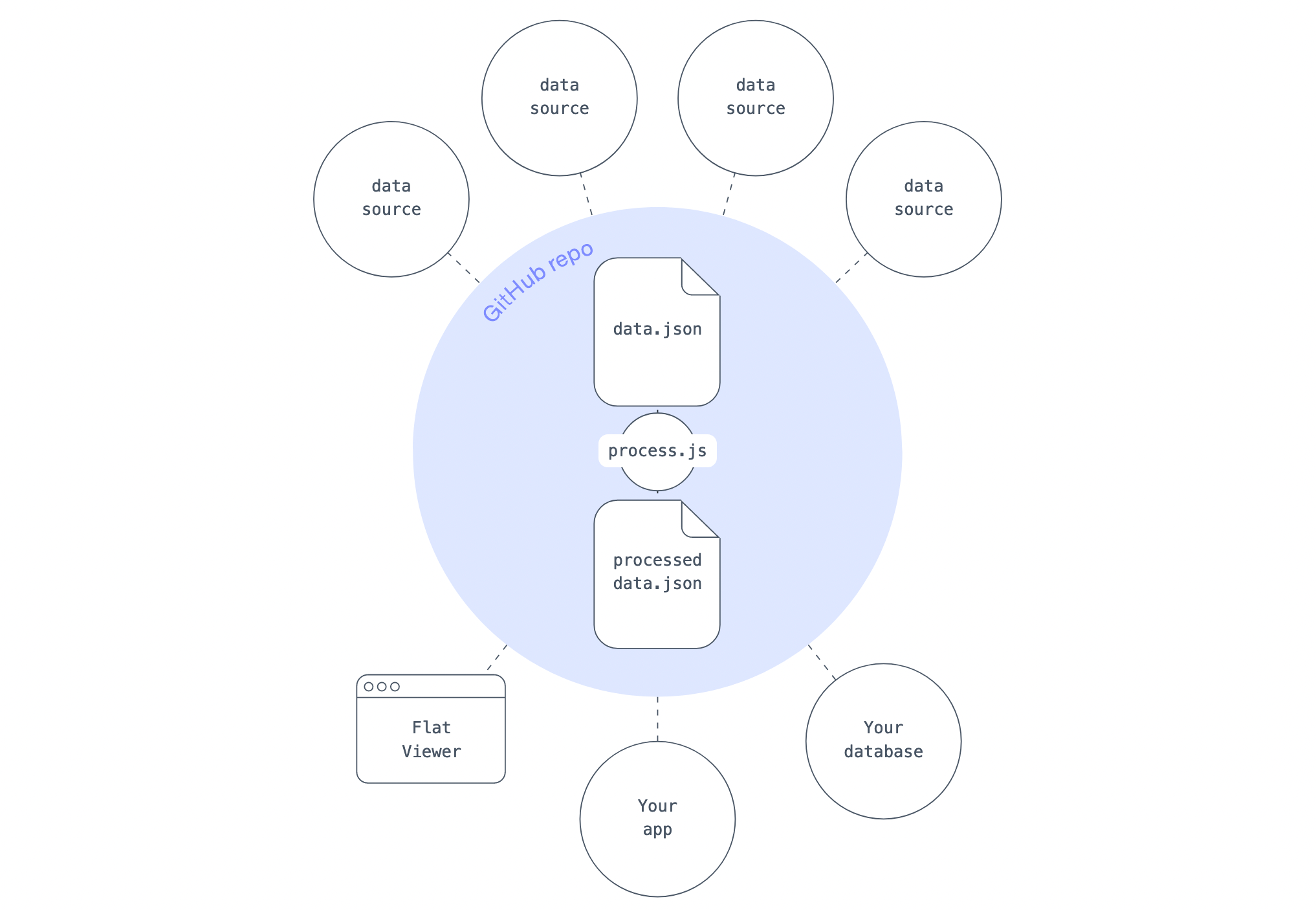
The idea, as seen above, is essentially to read in data (data.json), conduct some postprocessing (process.js), and output some better data (processed-data.json).
Doing it in R
The most essential step of a Flat Data project is postprocessing. This occurs after data retrieval and before data output, and it can be done in a few different languages. By default, the OCTO team's examples are done in JavaScript/TypeScript, and one user has given an example of postprocessing in Python. To the best of my knowledge, though, there aren't any examples of including R in the postprocessing stage, hence the reason for this post!
Using R in a Flat Data pipeline is as simple as installing the necessary packages and then sourcing your R cleaning script from a postprocessing TypeScript file. Let's explore how that works.
We'll be grabbing data from the Mapping Police Violence homepage, tidying it up, and then republishing it. (This cleaned data is the source for my visualization on police violence.) Here's a sample of the final data output:
01. Setup flat.yml
The first step in any Flat Data pipeline is to create .github/workflows/flat.yml, which will include the configuration for your project. You can do so by using GitHub's VSCode extension, or by creating your own YAML file manually. The YAML file we use in this project is remarkably similar to the boilerplate file, with a few differences:
name: Update data
on:
schedule:
- cron: 0 0 * * * # Runs daily. See https://crontab.cronhub.io/
workflow_dispatch: {}
push:
branches:
- main # Or master, or whatever branch you'd like to 'watch'
jobs:
scheduled:
runs-on: ubuntu-latest
steps:
# This step installs Deno, which is a Javascript runtime
- name: Setup deno
uses: denoland/setup-deno@main
with:
deno-version: v1.x
# Check out the repository so it can read the files inside of it and do other operations
- name: Check out repo
uses: actions/checkout@v2
# The Flat Action step. We fetch the data in the http_url and save it as downloaded_filename
- name: Fetch data
uses: githubocto/flat@v2
with:
http_url: https://mappingpoliceviolence.org/s/MPVDatasetDownload.xlsx # File to download
downloaded_filename: raw.xlsx # Name of downloaded file
postprocess: ./postprocess.ts # Script to run upon download completion
The tweaks you would make to this workflow are most likely in http_url and schedule. To confirm, visit GitHub's documentation.
02. Postprocess
We pick up at the last line of code in the previous chunk:
postprocess: ./postprocess.ts
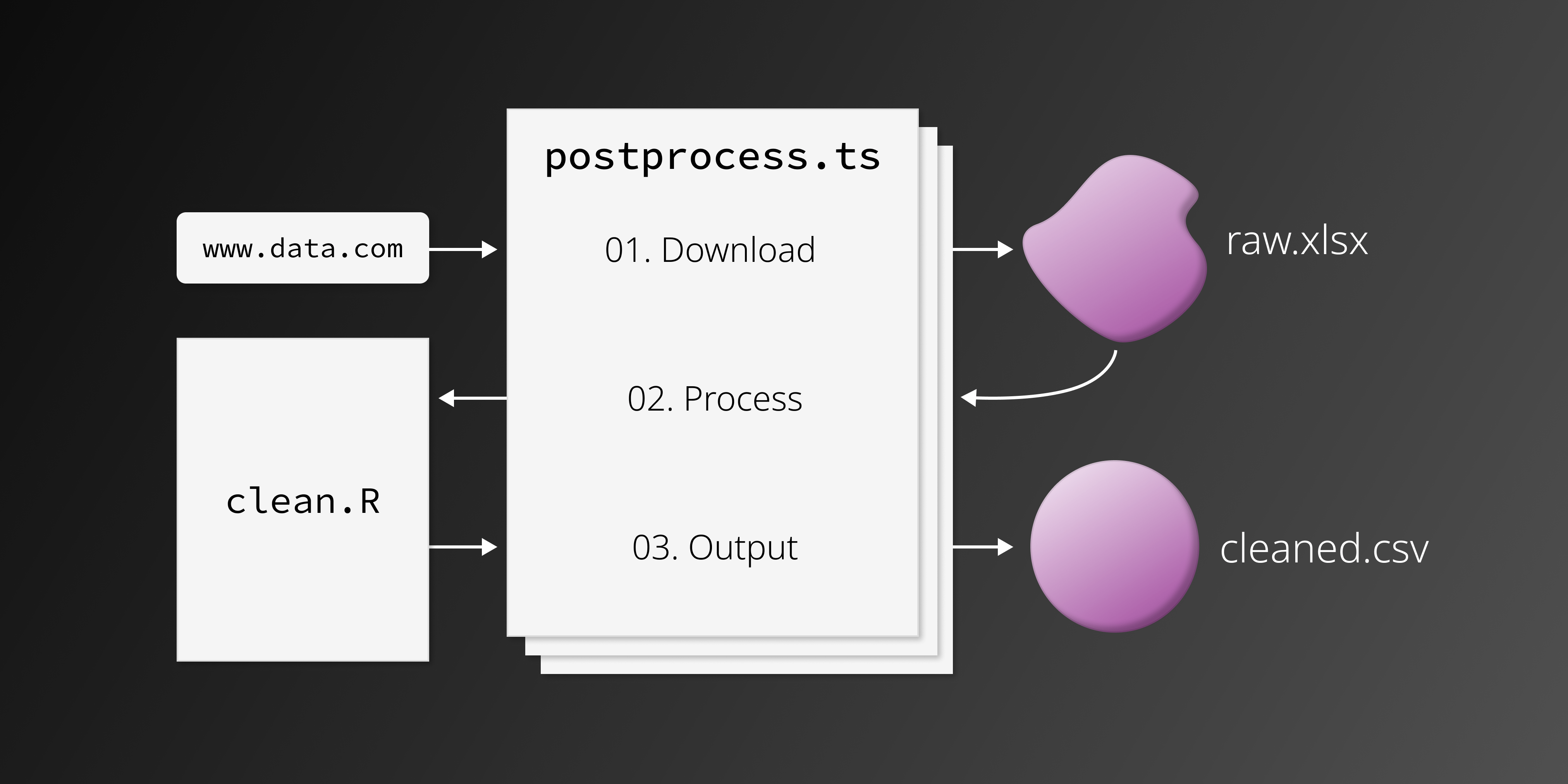
Here, we reference a TypeScript file titled postprocess.ts. Upon completion of the data download, GitHub will run this script for any additional processing steps. This file must be a .js or .ts file.
Those who are skilled in data wrangling with JavaScript might be able to write their additional processing in JavaScript itself, but few of us are skilled in data wrangling with JavaScript. Moreover, some users want to migrate their existing projects and workflows to Flat Data, and so including languages other than JavaScript (in this case, R) is essential.
The postprocess.ts file I use in my workflow looks like this (it might help to see how Deno works):
// 1. Install necessary packages
const r_install = Deno.run({
cmd: ['sudo', 'Rscript', '-e', "install.packages(c('dplyr', 'readxl', 'readr', 'lubridate', 'stringr'))"]
});
await r_install.status();
// 2. Forward the execution to the R script
const r_run = Deno.run({
cmd: ['Rscript', './clean.R']
});
await r_run.status();
The above script is rather simple: it 1) installs packages, and 2) runs the processing script, titled clean.R.
The first step is important. Package management was the biggest issue I ran into while setting up this workflow; if you're having issues, pay attention to this step. You'll need to identify all the packages that are required in your R processing script, but you can't install those packages in the script itself, due to virtual machine permissions. You instead have to run them via the command line, using sudo Rscript -e, as I do above (in step 1).
The command sudo Rscript -e precedes any regular function or command that you would run in an R script. It executes those commands via the command line, rather than within a script. (We add sudo to overcome system user permission problems.) For more, see this page.
03. Clean the data!
My clean.R script, which I reference at the bottom of postprocess.ts looks like this:
# Load libraries
library(dplyr)
library(stringr)
# Read in data, with the same name that we specified in `flat.yml`
raw_data <- readxl::read_excel("./raw.xlsx")
# All the processing!
clean_data <- raw_data %>%
rename("Date" = `Date of Incident (month/day/year)`,
"Link" = `Link to news article or photo of official document`,
"Armed Status" = `Armed/Unarmed Status`,
"Age" = `Victim's age` ,
"Race" = `Victim's race`,
"Sex" = `Victim's gender`,
"Image" = `URL of image of victim`,
"Name" = `Victim's name`) %>%
mutate(Zipcode = as.character(Zipcode),
Year = lubridate::year(Date),
Sex = ifelse(is.na(Sex), 'Unknown', Sex)) %>%
arrange(Date)
### Additional processing goes here...
# Output data
readr::write_csv(clean_data, "./output.csv")
Obviously, the content in the above cleaning script is irrelevant. It functions as any other R script would: it reads in data (based on the data we downloaded in postprocess.ts), does some cleaning, and then outputs the new data. The real script is around 55 lines. Now you know why keeping the postprocessing in R was preferable!
In Sum
Upon completing these steps and pushing the above to a repository, GitHub will automatically set up the action and run it on a daily basis. You can then examine the logs for each run in the Actions tab. This tab will be helpful for debugging, and you can force workflow executions manually here as well. In sum, the process of carrying out a GitHub Flat Data workflow, with the addition of an R postprocessing script, looks something like this:
Thanks for reading! You might learn more by perusing the GitHub repository that accompanies this post; otherwise, please send any questions via Twitter 🙂